Around the world, many people are doing their best to combat the current Covid-19 pandemic in many different (and creative) ways: from 3D printed face shields and homebrew face masks to replacements for full-fledged mechanical ventilators, the outpouring of ideas and contributions has been impressive and motivating. At universities, researchers try to model and understand the coronavirus, a challenging task that involves complex calculations and requires massive computing power, and that has also been receiving help from volunteers.
In this regard, the best-known project is Folding@home (F@h or FAH). Its website provides a program that anyone can run on their computer to contribute to research by donating their household’s unused computing power. The program downloads a task, works on it and sends the result back, contributing to the bigger task, which is modeling the virus.
A little about the project
Folding@home was created in 2000 by the Pande Lab at the Stanford University, led by professor Dr. Vijay Pande. He sought to understand how proteins self-organize (a process called protein folding, hence the project name) and why this process sometimes goes wrong, causing issues such as cancer and Alzheimer’s disease. Since 2018, the project is based at the Washington University in St. Louis, led by professor Dr. Gregory Bowman, a former student of Dr. Pande, and receives contributions from other universities and companies.
The project has already produced big findings, reported in 224 research papers, which include work on antibiotic-resistant bacteria and proteins from the Ebola virus.
By January 2020, Folding@home had the support of 30,000 users, which was already pretty good, but that number suddenly jumped to 400,000 in March, due to people’s interest in helping find a therapy for Covid-19. Many of those new users embarked on the project together with NVIDIA, which tweeted out a “call to arms”.
PC Gamers, let’s put those GPUs to work.
— NVIDIA GeForce (@NVIDIAGeForce) March 13, 2020
Join us and our friends at @OfficialPCMR in supporting folding@home and donating unused GPU computing power to fight against COVID-19!
Learn more → https://t.co/EQE4u7xTZT pic.twitter.com/uO0ZCq8PEv
Folding@home reached a peak performance of 1.5 exaFLOPs, making it more than seven times faster than the world’s current fastest supercomputer, IBM Summit.
Minimum system requirements
There are no minimum hardware requirements to enter Folding@home, all computers can contribute to the project. However, if the computer is too slow, it might not be fast enough to make the deadlines of typical tasks. The project estimates that computers built in the last 5 years or so can well meet the demand. On the other side, gaming PCs with a NVIDIA or AMD graphics card are the ideal candidates due to their greater capacity for parallel processing.
Note that tasks are very processor-intensive and your computer might overheat.
Is this program open source?
Maybe this is important to you, so I think it’s worth noting: the Folding@home program is not open source, although it is built from several open source tools, among which are mentioned on the project’s website: Gromacs, TINKER, Amber and MPICH. The project states that any changes it makes to those tools are contributed back.
The project’s justification for not open sourcing the program is the concern for scientific integrity, since malicious people could modify the code to produce bogus scientific results, which would make the whole project pointless.
I won’t discuss whether I agree or not, I just reproduced the information I found on the project’s website.
If despite of that you want to donate your computing power for Covid-19 research, now you are going to see how. As usual, I focus on openSUSE and Linux Kamarada. In other Linux distributions, the step-by-step may be slightly different.
Download
Go to the Folding@home download page:
Observe that Folding@home does not officially support openSUSE, but we can make it work. See that there are 3 RPM packages for close distributions (RedHat, CentOS and Fedora):
You don’t need to actually install all the 3 packages. See what each one is:
- FAHClient: this is the most important package because it contains:
- the client software that downloads tasks, works on them and sends the results back, typically runs behind the scenes (in background), as a system service, without user interaction (back-end)
- a simple web interface which you can access from the web browser to monitor the client and minimally set it up (front-end)
- FAHControl: a graphical user interface (GUI) application that allows you to monitor the client and make more advanced settings, this package is optional
- FAHViewer: a GUI application that allows you to view the model on which your computer is working, this package is optional
For demonstration purposes, I’m going to download and install each of them. Feel free to download and install just the one(s) you want.
FAHClient
After downloading the fahclient RPM package, open a terminal window in the folder where it was downloaded and install it by running this command as administrator (root user):
1
# zypper in fahclient-7.6.9-1.x86_64.rpm
The zypper package manager informs that it cannot find a dependency of the package:
1
2
3
4
5
6
7
8
9
Loading repository data...
Reading installed packages...
Resolving package dependencies...
Problem: nothing provides bzip2-libs needed by fahclient-7.6.9-1.x86_64
Solution 1: do not install fahclient-7.6.9-1.x86_64
Solution 2: break fahclient-7.6.9-1.x86_64 by ignoring some of its dependencies
Choose from above solutions by number or cancel [1/2/c/d/?] (c):
Remember that this RPM package was not made for openSUSE, which provides the bzip2 libraries in a package named libbz2-1, which is already installed by default. Thus, type 2 (meaning you want to ignore the missing dependency) and hit Enter:
1
2
3
4
5
6
7
8
9
10
Choose from above solutions by number or cancel [1/2/c/d/?] (c): 2
Resolving dependencies...
Resolving package dependencies...
The following NEW package is going to be installed:
fahclient
1 new package to install.
Overall download size: 3.5 MiB. Already cached: 0 B. After the operation, additional 8.9 MiB will be used.
Continue? [y/n/v/...? shows all options] (y):
Hit Enter to accept the default option (y, which here means “yes, continue”).
Then, zypper is faced with another problem. The package is not signed and, therefore, it’s not possible to do authenticity verification:
1
2
3
4
5
6
Retrieving package fahclient-7.6.9-1.x86_64 (1/1), 3.5 MiB ( 8.9 MiB unpacked)
fahclient-7.6.9-1.x86_64.rpm:
Package is not signed!
fahclient-7.6.9-1.x86_64 (Plain RPM files cache): Signature verification failed [6-File is unsigned]
Abort, retry, ignore? [a/r/i] (a):
This error message is common when installing a manually downloaded RPM package (not a package retrieved from a known repository). You can safely type i (ignore) and press Enter:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
Abort, retry, ignore? [a/r/i] (a): i
Checking for file conflicts: .......................................................................[done]
(1/1) Installing: fahclient-7.6.9-1.x86_64 .........................................................[done]
Additional rpm output:
Unknown option: levels
usage:
chkconfig -A|--allservices (together with -l: show all services)
chkconfig -t|--terse [names] (shows the links)
chkconfig -e|--edit [names] (configure services)
chkconfig -s|--set [name state]... (configure services)
chkconfig -l|--list [--deps] [names] (shows the links)
chkconfig -L|--liston [--deps] [names] (as -l, enabled in at least 1 level)
chkconfig -c|--check name [state] (check state)
chkconfig -a|--add [names] (runs insserv)
chkconfig -d|--del [names] (runs insserv -r)
chkconfig -h|--help (print usage)
chkconfig -f|--force ... (call insserv with -f)
chkconfig [name] same as chkconfig -t
chkconfig name state... same as chkconfig -s name state
chkconfig --root=<root> ... use <root> as the root file system
Starting fahclient ... FAIL
zypper finishes the installation, but shows a few more error messages. Again, remember that this RPM package was not made for openSUSE. Although one of those error messages informs that the client failed to start, that’s not the case.
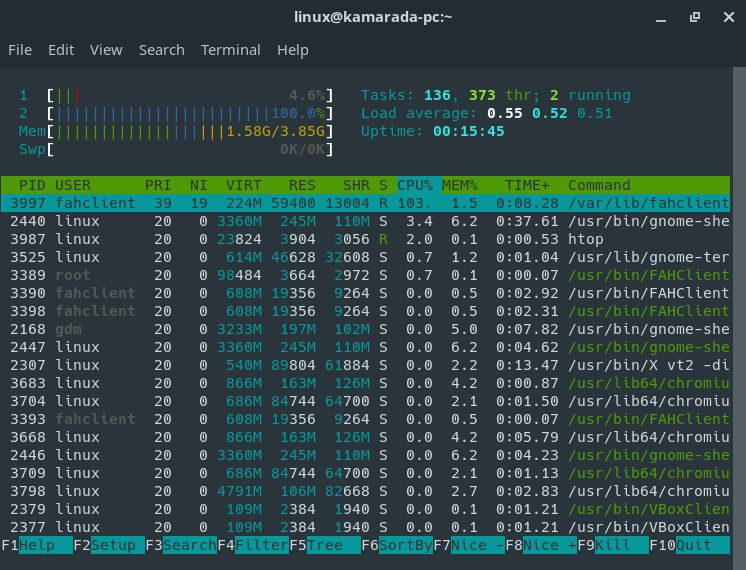
If you use the htop command, you will notice that not only the client is running, but it also quickly puts your processor to work hard, reaching 100% usage:
1
$ htop
To exit htop, hit F10 or click Quit on the bottom-right corner of the window.
Reload the system services (needed because FAHClient is still based on /etc/init.d):
1
# systemctl daemon-reload
And enable the FAHClient service so that it gets started automatically at boot time:
1
# systemctl enable FAHClient
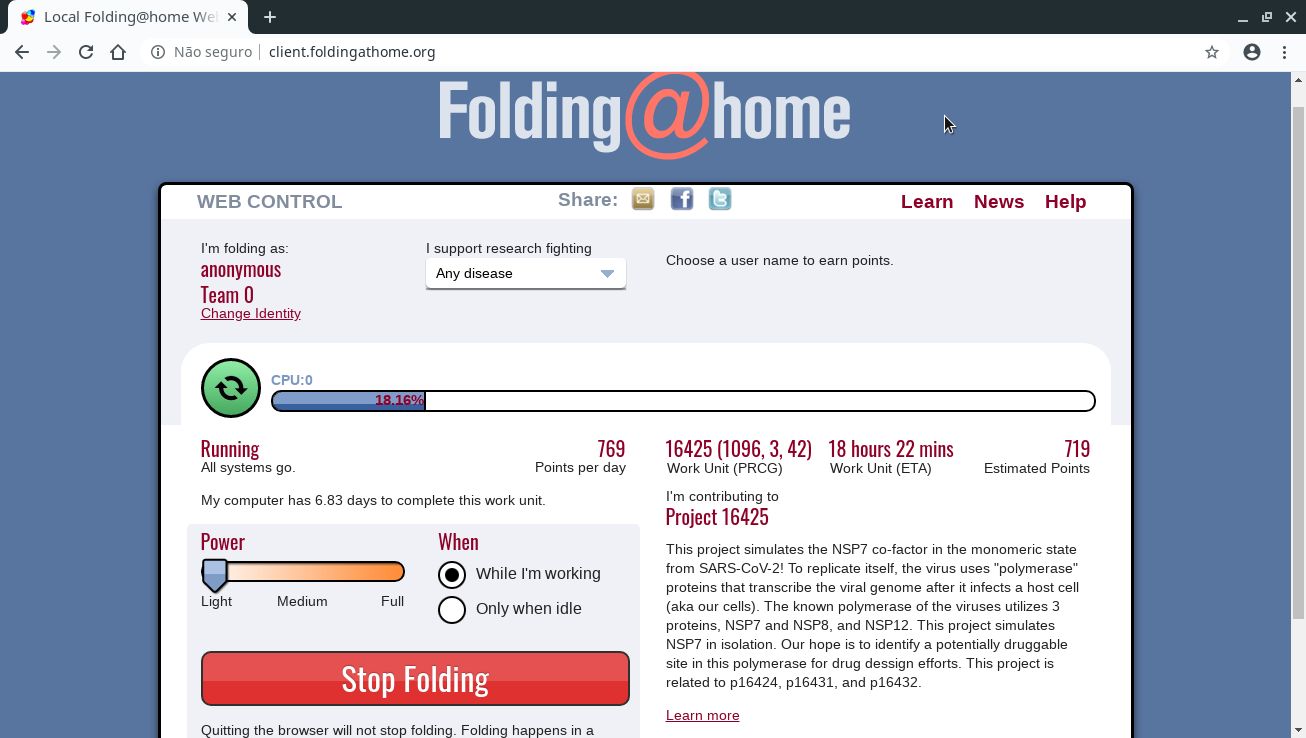
To finish the configuration, open your web browser and go to client.foldingathome.org:
That page redirects to your FAHClient’s web interface. If you prefer, you can access it directly at localhost:7396.
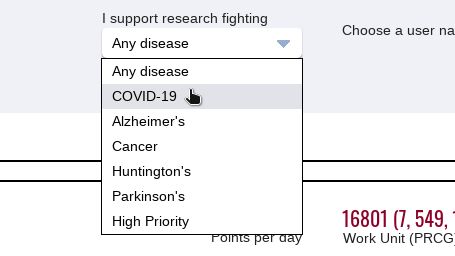
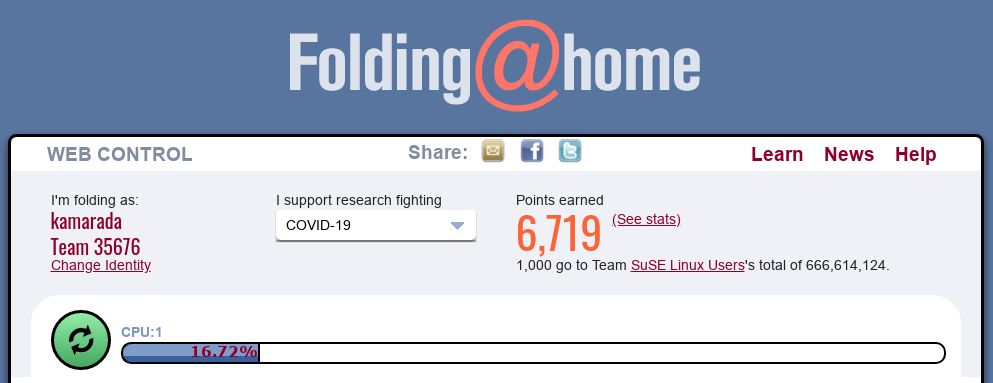
In I support research fighting, there is a dropdown menu that you can open and select a disease to direct your computer efforts:
You can choose a line of research of your choice (e.g. COVID-19) or leave the default, which is to contribute to researches against Any disease.
In Power, you can define how much of your processing power will be donated (default is Light).
In When, you can define when the computer will work on protein folding: during all the time, even while you are using the computer (While I’m working, default) or only while you’re away from the computer (Only when idle).
If you need, you can stop the FAHClient’s work at any time by clicking the Stop Folding button.
Note that closing this browser tab does not stop FAHClient’s work, which runs in background.
That’s it, congratulations! You are already contributing to researches. If you want, you can stop here, the following steps are optional. Follow me the curious ones!
Next, I added some lines talking about identity and teams (optional).
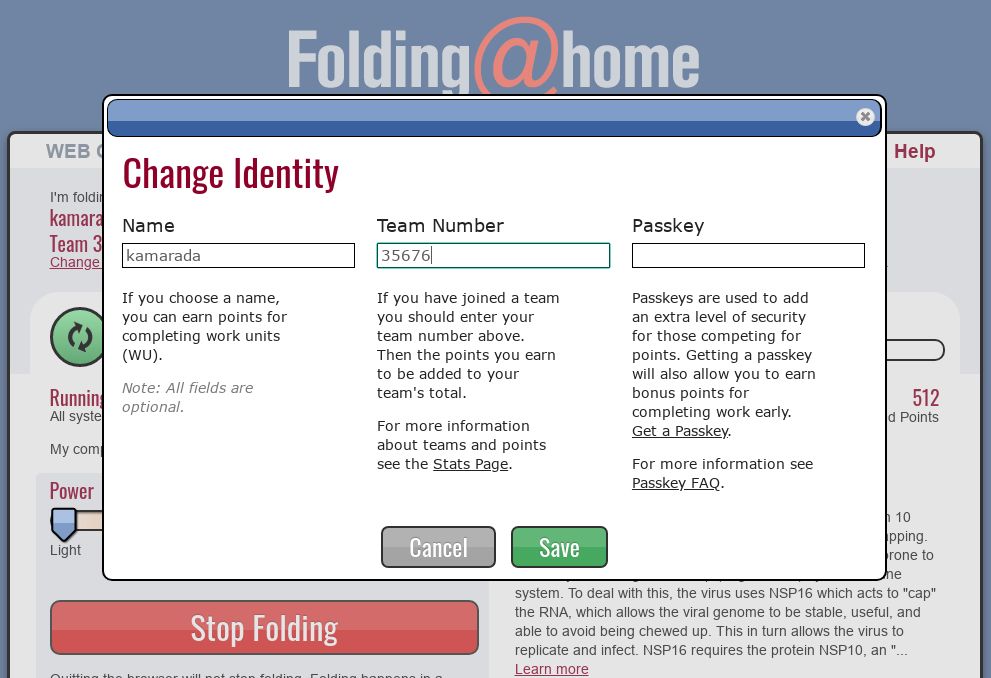
By default, you contribute to the project anonymously, you don’t need to register. But, optionally, you can choose a username for yourself, as well as indicate that you belong to a team. If you want to do this, in your FAHClient’s web interface, click the Change Identity link.
Fill in the Name and the Team Number fields with a username — it’s recommended that you stick to just letters, numbers and underscore — and the number of the team you have joined. Examples of teams include:
- openSUSE Users team #35676 (at the time of writing, position 389 in the team ranking, among almost 253 thousand registered teams)
- Team SUSE #12841 (position 212 in the team ranking)
Now your name and team appear on the web interface and you start earning points for you and your team as your computer advances in tasks:
Maybe these points cannot be exchanged for any concrete reward, but they encourage competition between teams to see which one contributes the most to the project.
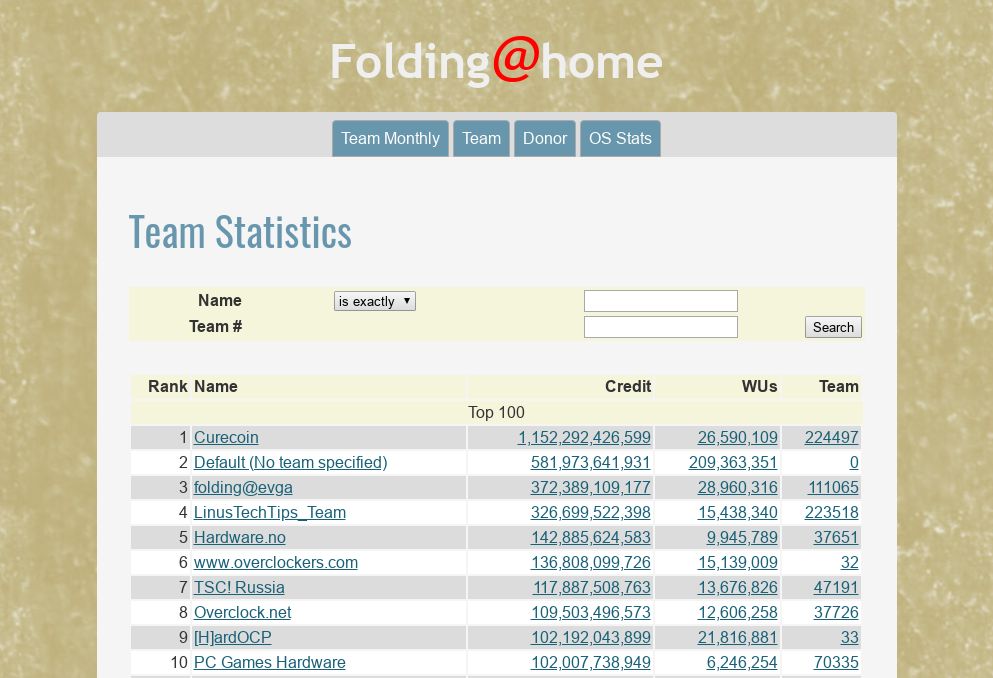
The searchable team ranking is available on the project’s Stats page:
It caught my attention that Petrobras (a Brazilian oil company) has a team, which is 59th in the team ranking. Googling, I found out that two of the company’s high performance computers are contributing to Folding@home. Points to Brazil!
People of the openSUSE project and the SUSE company are committed to helping the Folding@home project and combating Covid-19. If you want more information, see:
- Folding@home - openSUSE Wiki
- SUSE’s Commitment to Combat COVID-19 - SUSE Blog
- Folding@home with openSUSE to assist with COVID-19 - openSUSE - reddit
- TUMBLEWEED FAH ( Folding@Home) packages for TW - openSUSE Forums
FAHControl
You can install the fahcontrol package the same way:
1
# zypper in fahcontrol-7.6.9-1.noarch.rpm
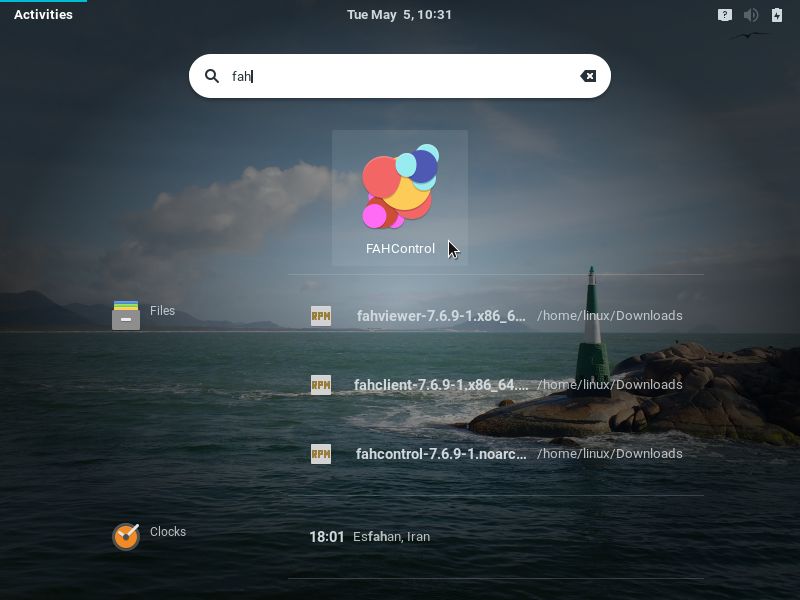
Once installed, to start FAHControl, if you use the GNOME desktop, click Activities, on the top-left screen corner, type fah and click the FAHControl icon:
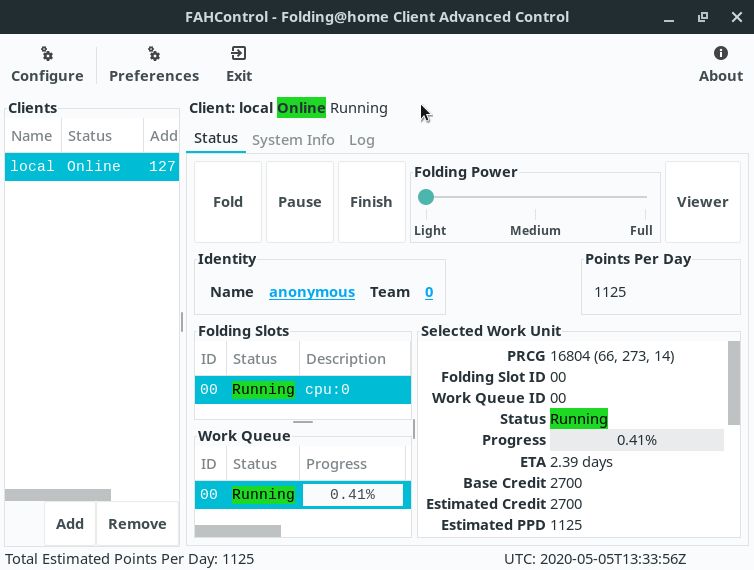
Note that this application presents the same options of the web interface and more:
If you need, you can pause the FAHClient’s work at any time by clicking the Pause button, or abort it completely using the Finish button. In both cases, to resume working, click Fold.
Note that closing this program does not stop FAHClient’s work as well.
FAHViewer
You can install the fahviewer package the same way:
1
# zypper in fahviewer-7.6.9-1.noarch.rpm
Once installed, to start FAHViewer, if you use the GNOME desktop, click Activities, type fah and click the FAHViewer icon:
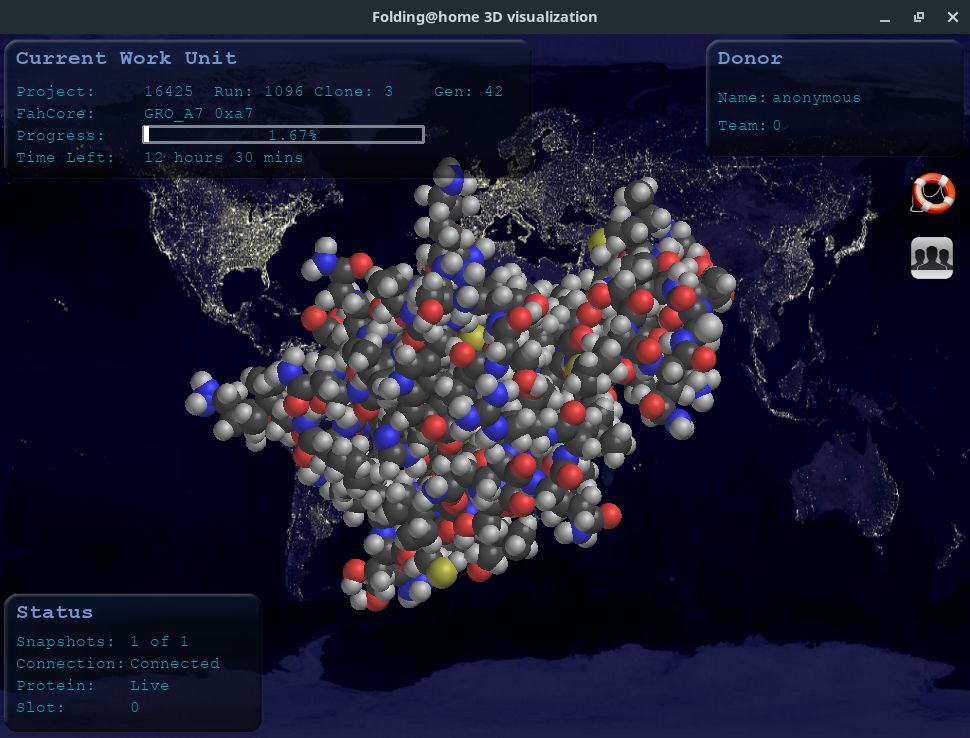
This app shows you graphically the model your computer is working on:
If you want, you can rotate the protein 3D image using the mouse.
References
- Folding@Home: How Your PC Can Help in the Fight Against COVID-19
- Folding@home - Fighting disease with a world wide distributed super computer
- Folding@home - Wikipedia
- So What Is Protein Folding, Anyway? - Hackaday
- The coronavirus pandemic turned Folding@Home into an exaFLOP supercomputer - Ars Technica
- init.d - What’s the easiest way to make my old init script work in systemd? - Server Fault